 |
 |
 |
| |
Table 2-3. Summary of the ChIP-seq data. (hg19)
|
| |
21% |
1% |
|
| IP total |
WCE total |
Peak |
IP total |
WCE total |
Peak |
|
| DLD-1 |
Pol II |
14,397,292 |
19,133,214 |
12,405 |
16,431,596 |
17,512,096 |
17,512,096 |
| H3ac |
11,545,404 |
11,430,778 |
16,869 |
12,406,376 |
11,049,033 |
21,414 |
| H3K4me3 |
15,126,753 |
19,803,256 |
27,197 |
14,673,411 |
15,244,721 |
29,693 |
| H3K27me3 |
9,796,488 |
15,762,543 |
1,434 |
8,230,411 |
14,789,992 |
1,562 |
|
| MCF7 |
Pol II |
9,744,263 |
20,224,021 |
25,188 |
9,709,791 |
12,724,278 |
19,311 |
| HIF1A |
25,763,246 |
18,197,242 |
94,085 |
19,390,075 |
17,154,571 |
107,589 |
|
| HEK293 |
Pol II |
10,086,952 |
5,772,027 |
27,895 |
- |
- |
- |
|
| TIG3 |
Pol II |
20,068,484 |
16,048,237 |
8,525 |
9,078,433 |
8,178,309 |
7,443 |
| H3ac |
11,610,062 |
14,092,706 |
10,818 |
10,541,222 |
13,273,725 |
7,941 |
| H3K4me3 |
10,177,529 |
19,487,961 |
31,549 |
9,438,915 |
16,891,626 |
25,720 |
| H3K27me3 |
12,207,120 |
14,020,038 |
9,035 |
11,616,140 |
17,176,075 |
14,968 |
| HIF1A |
18,092,915 |
14,732,632 |
13,782 |
16,467,590 |
15,541,811 |
674 |
|
| |
IL4+ |
IL4- |
|
| IP total |
WCE total |
Peak |
IP total |
WCE total |
Peak |
|
| Ramos |
Pol II |
7,192,077 |
2,786,371 |
17,426 |
8,605,893 |
3,299,301 |
19,233 |
| H3ac |
31,342,754 |
27,241,289 |
51,325 |
37,442,045 |
27,794,747 |
35,708 |
| H3K4me3 |
36,933,113 |
24,593,765 |
27,347 |
36,933,113 |
24,593,765 |
26,105 |
| STAT6 |
4,684,426 |
5,008,189 |
600 |
4,550,900 |
4,842,993 |
73 |
|
| Beas2B |
Pol II |
6,869,914 |
3,188,421 |
13,453 |
7,911,582 |
3,724,882 |
8,171 |
| H3ac |
35,939,360 |
26,678,038 |
37,462 |
32,576,988 |
29,388,072 |
32,767 |
| H3K4me3 |
33,520,360 |
27,485,630 |
24,264 |
29,914,406 |
26,630,062 |
23,466 |
| STAT6 |
4,886,971 |
3,188,421 |
772 |
7,154,872 |
3,724,882 |
314 |
|
|
|
 |
 |
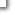 |
|