 |
 |
 |
| |
| A. | B. |
 |
 |
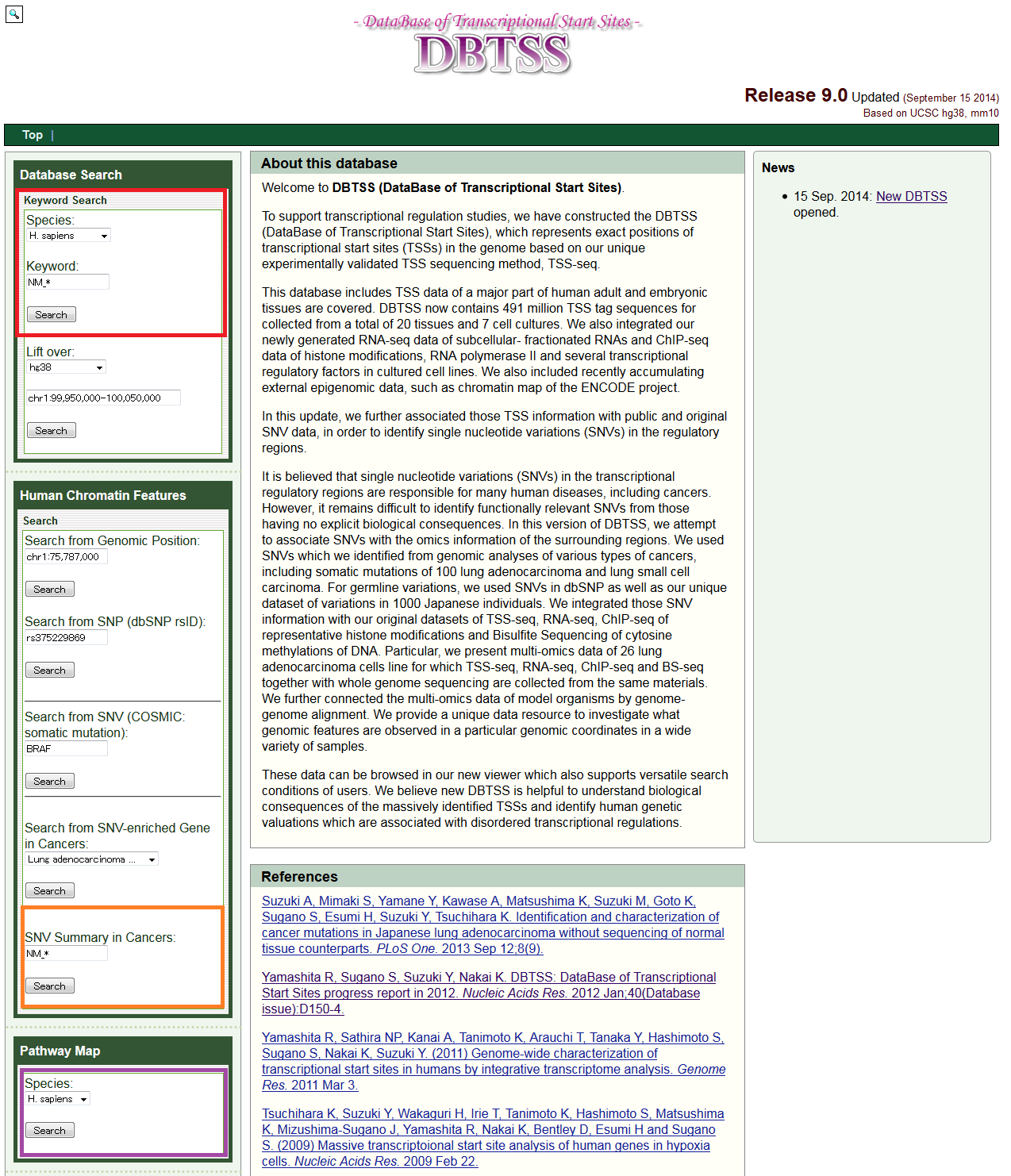
- Top page of DBTSS. A simple search for "TSS Viewer" and "Genome Viewer" can be made by specifying a keyword, such as a gene name 'BRAF' in the Database Search at the left frame (red box). Search by "SNV Summary in Cancers" and "Pathway Maps" can be made from the positions indicated by orange and purple boxes, respectively.
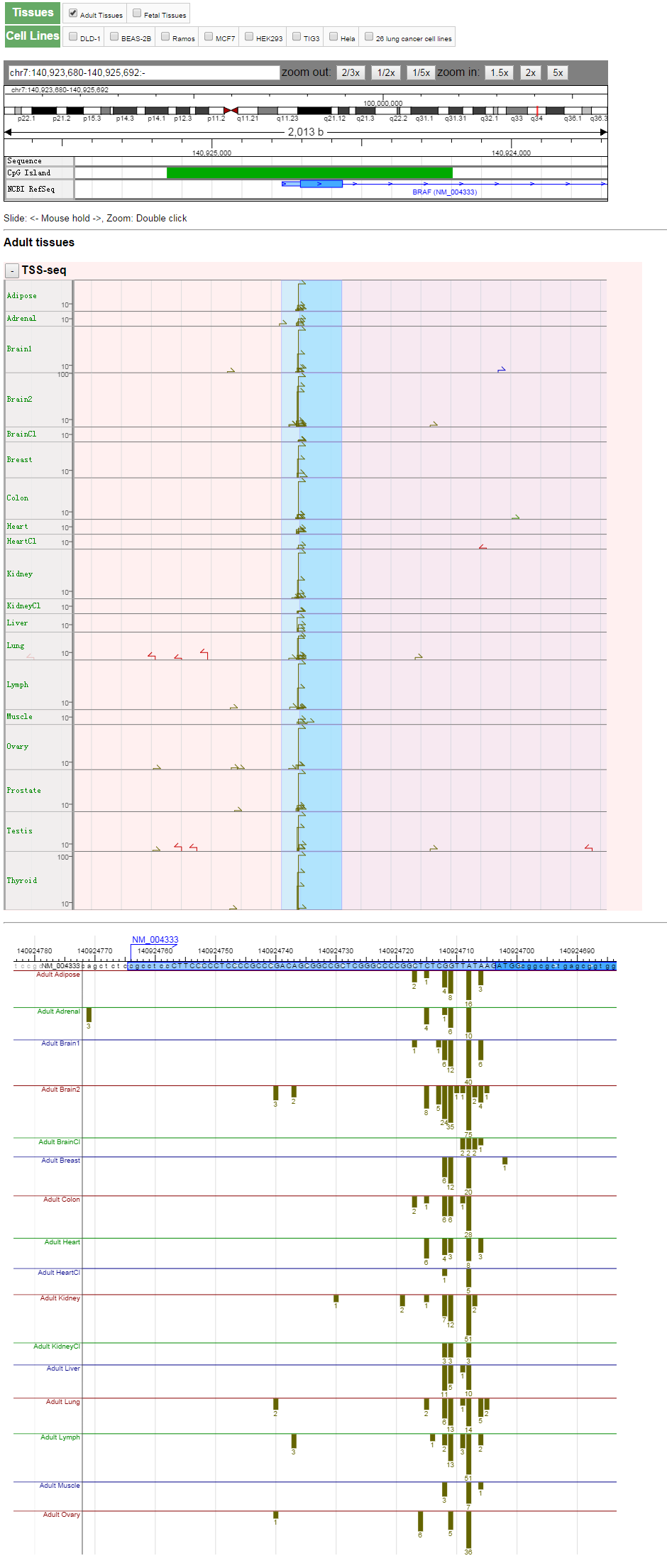
- A part of the TSS Viewer display for the BRAF gene. The overview and the detailed positions of the TSSs are shown in the upper and lower panels, respectively. Many of the fields are expandable.
|
| C. | D. |
 |
 |
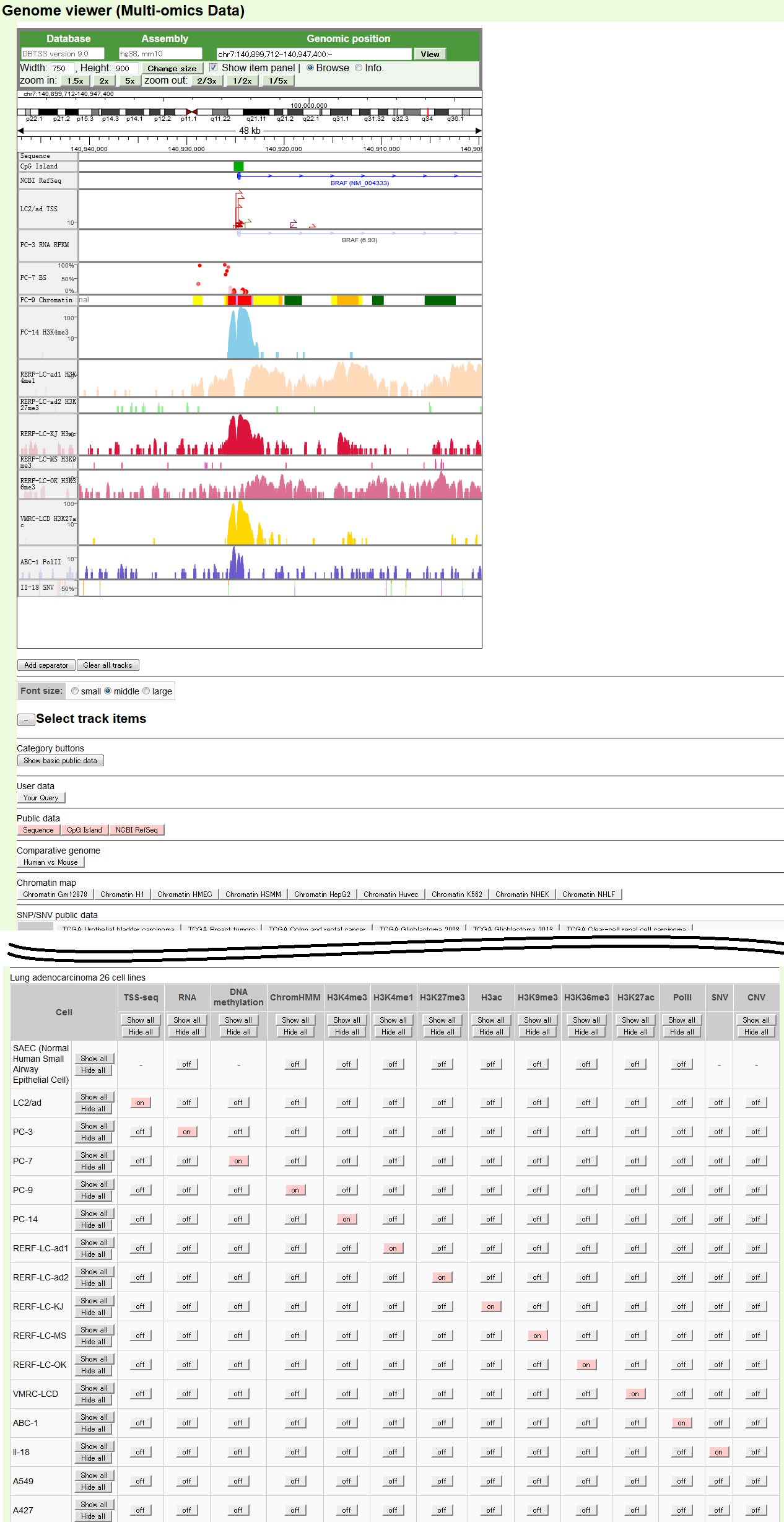
- The default display of Genome Viewer for the BRAF gene. Displayed items are as indicated in the margin. The displayed items can be controlled from the panels located under the "Select track items" headline.
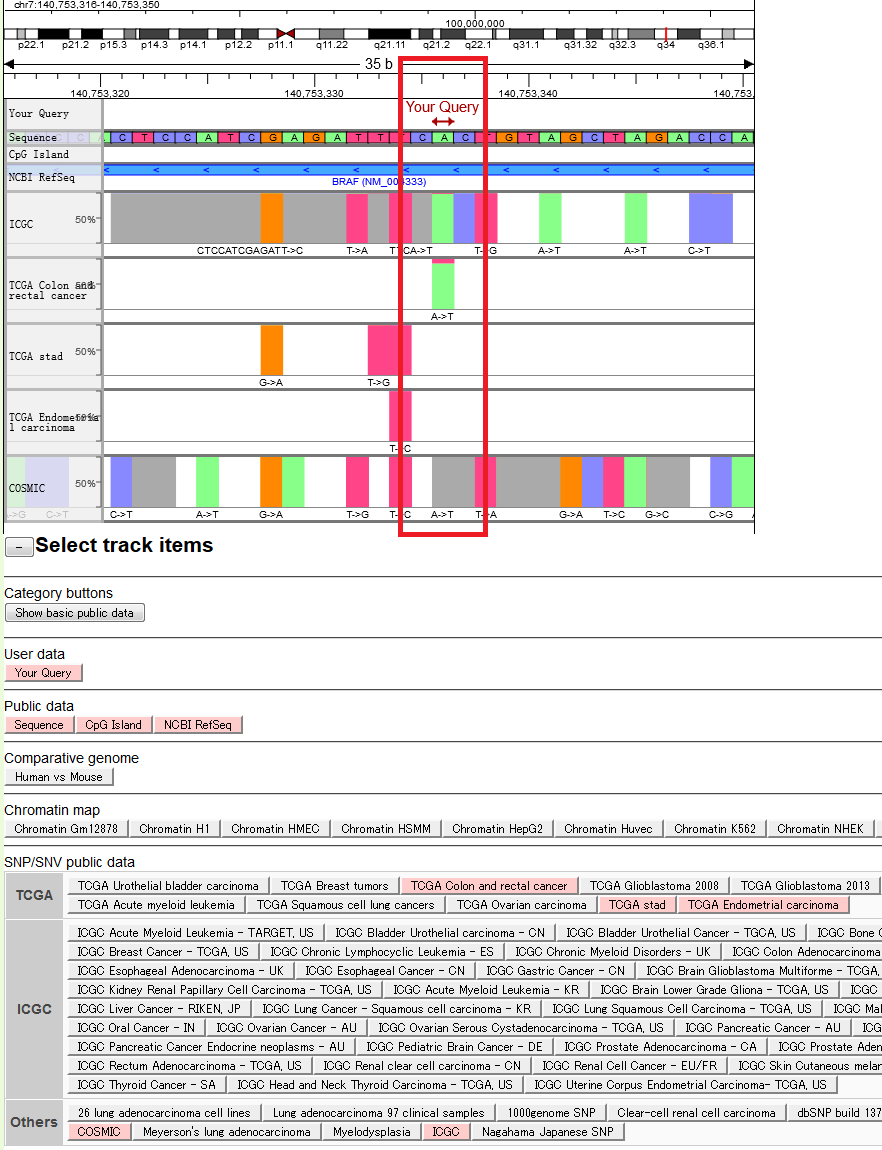
- A sample output of SNV information for the BRAF gene. Surrounding region of a previously reported cancer driver mutation (V600E of the BRAF gene; highlighted in red box), is displayed.
|
| E. | F. |
 |
 |
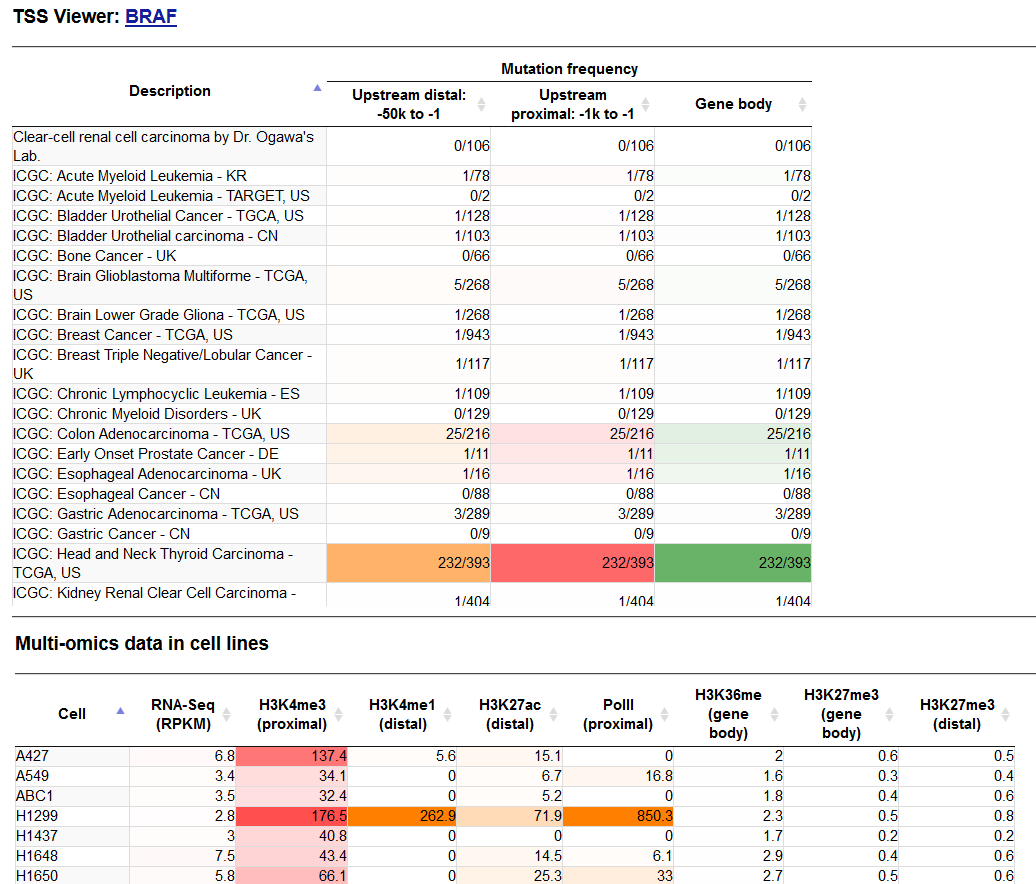
- Upper panel: A part of the Mutation frequency table for the BRAF gene. Enriched fields are as highlighted; lower panel: Summary of multi-omics data mainly collected from cell lines.
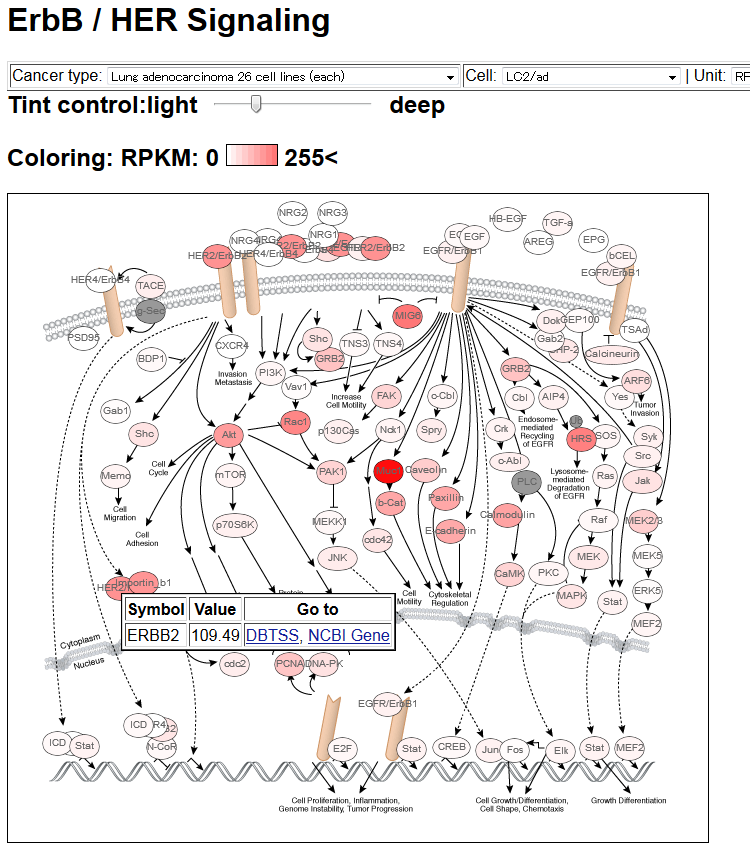
- Pathway Map representation of characteristic genes. In this example, gene expression level (in RPKM) of node genes in a lung adenocarcinoma cell line, LC2/ad, in the ErbB/HER signaling pathway is shown. Further links will appear when the users click the circle corresponding to each gene.
|
|
|
 |
 |
 |
|