 |
 |
 |
 |
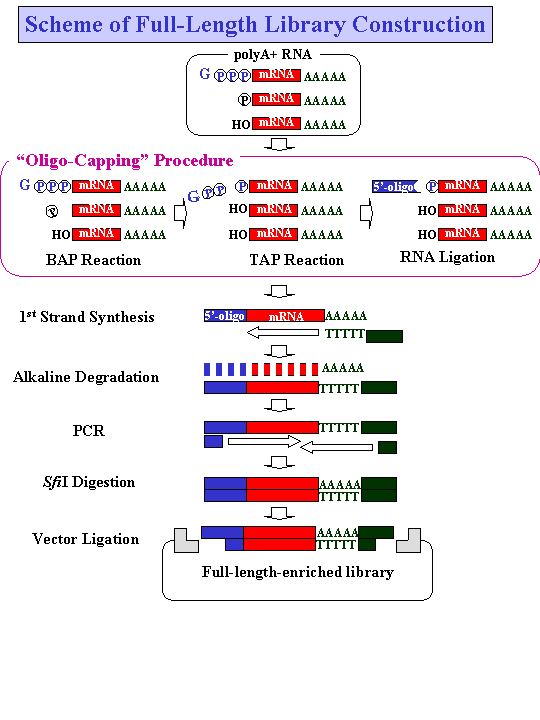
!HOligo-Capped!I cDNA libraries were constructed according
to the scheme shown. The cap structure of the mRNA was replaced
with the 5!G-oligo by the !HOligo-Capping!I method, which consists
of three enzymatic reaction steps. Bacterial alkaline phosphatase
(BAP) hydrolyses the phosphate of truncated mRNA 5!G-ends whose
cap structures have been broken down. Tobacco acid pyrophosphatase
(TAP) removes the cap structure, leaving the phosphate at the 5!G-end.
T4 RNA ligase, which requires a phosphate at the 5!G-end as its
substrate, selectively ligates the 5!G-oligo to the 5!G-end that
originally had the cap structure. Using !HOligo-Capped!I mRNA, first
strand cDNA was synthesized with dT adapter primer. After alkaline
degradation of the RNA, first strand cDNA was amplified by PCR,
digested with restriction enzyme SfiI and cloned into a plasmid
vector. For further details of the procedure, see reference 1 and
2. RNA and DNA molecules are represented by solid lines, the 5!G-oligo
by gray boxes, and PCR primers by shaded boxes. Gppp: cap structure;
p: phosphate; OH: hydroxyl
|
 |
 |
 |
 |
|